Introduction
Automation of library preparation is very important to reduce opportunities for error, increase reproducibility, and reduce the amount of hands-on time allowing researchers to generate sequence data from DNA more efficiently. By utilizing the CyBi®-FeliX, a benchtop instrument for a variety of automated liquid handling tasks, the HaloPlex Target Enrichment System Kit for NGS library preparation for Ion Torrent™ Sequencing was automated to achieve reliable final prepared libraries with high accuracy for downstream sequencing. This application note describes the successful proof of principle to generate high-quality DNA libraries on CyBi®-FeliX for personal genome analysis with the Ion Torrent™ Sequencing System.
Analytik Jena’s liquid handling portfolio includes CyBi®-FeliX (Figure 1), a compact and flexible liquid handling platform for a variety of automated liquid handling tasks. The system has 12 deck positions on two levels for microplates, tube racks, reservoirs and tips on smallest footprint, including accessories like gripper, magnet adapter and thermal shaker.
Library preparation is a critical, hands-on and time-consuming step in the Next Generation Sequencing (NGS) workflow. To simplify the NGS library preparation process Analytik Jena developed a protocol for the HaloPlex Target Enrichment System Kit with minimal user intervention to prepare 8 library samples for multiplexed sequencing on the Ion Torrent™ platform.
The Institute for Laboratory Medicine in the Donauspital in Vienna was testing the CyBi®-FeliX liquid handling platform to prepare DNA sample libraries from patient samples. Target enrichment of genomes greatly enhances the efficiency of Next Generation Sequencing, allowing researchers to focus on specific regions of interest. The Kit is suited to deep sequencing of small panels of genes (up to 2.5 Mb), such as those required for studies of inherited disorders, cancer, or infectious and neurologic diseases.
Material and methods
Kits and reagents
- Library preparation for Ion Torrent™ Sequencing was performed using 8 normalized patient DNA samples (225 ng), isolated with the innuPREP Blood DNA Mini Kit (Analytik Jena AG). Patient DNA was quantified using the Qubit fluorometer (Life Technologies). For each sample (patient DNA) to be sequenced, an individual target-enriched, barcoded library was prepared. The HaloPlex probes were designed using Agilent´s SureDesign tool.
- HaloPlex Target Enrichment System Kit (Agilent)
- HaloPlex Custom Panel Tier 1, ION (G9902B)
- Agencourt AMPure XP Kit 5 ml / 60 ml (Beckman Coulter Genomics p/n A63880, p/n A63881)
- Herculase II Fusion Enzyme with dNTPs (100 mM; 25 mM for each nucleotide) (Agilent p/n 600677)
- Nuclease-free Water (not DEPC-treated) (Ambion Cat #AM9930)
- 10 M NaOH, molecular biology grade (Sigma, p/n 72068)
- 2 M acetic acid (Sigma, p/n 72068)
- 10 mM Tris-EDTA, pH 8.0
- 100% Ethanol, molecular biology grade (Sigma-Aldrich p/n E7023)
- InnuPREP Blood DNA Mini Kit (Analytik Jena AG, 845-KS�1020050)
- Quant-iT dsDNA BR Assay Kit, for use with the Qubit fluorometer (100 assays, 2-1000 ng, 500 assays, 2-1000 ng) (Life Technologies p/n Q32850, Life Technologies p/n Q32853)
Consumables
- Hard shell 96-well PCR plate (Eppendorf twin.tec, skirted, 0030 128.680)
- 8-well PCR strip tubes with caps (Nippon Genetics p/n FG�088WF)
- 2 ml tubes (STARLAB, E1420-2340)
- Skirted 0.5 ml tubes (STARLAB, E1405-2120)
- Tube Caps (STARLAB, 1480-0199)
- CyBi®-TipBox 96/250 µl (OL 3811-25-637-S)
- CyBi®-TipBox 96/50 µl (OL 3811-25-535-N)
- Qubit assay tubes (Life Technologies p/n Q32856)
Equipment
- CyBi®-FeliX (OL 5015-24-100) with CyBio® Composer Software and CyBi®-FeliX Head R 96/250 µl (OL 3316-14-850)
- 8-channel adapter Head R 96 (OL 3317-11-330)
- 12-channel adapter Head R 96 (OL 3317-11-340)
- Gripper (OL 3317-11-800)
- Tube Rack 2.0 ml for 24 tubes (844-00136-0)
- IsoFreeze PCR Rack SBS (LTF0045)
- Magnet Adapter M96 (Biovendis, 209601)
- BioShake 3000-T elm with PCR plate adapter (QInstruments, 1808-0517)
- Mounting Kit BioShake 3000 Series (OL 3317-23-690)
- Waste box (844-00430-0)
- Reagent tube Support (OL 3317-11-240) with Reagent tube 30 ml (2320230-00)
- Thermal cycler
- 2200 TapeStation (Agilent p/n G2964AA or G2965AA)
- High Sensitivity D1000 ScreenTape (Agilent p/n 5067-5584)
- High Sensitivity D1000 Reagents (Agilent p/n 5067-5585)
- Qubit 2.0 Fluorometer (Life Technologies p/n Q32866)
- Ion PGM sequencer (Life Technologies)
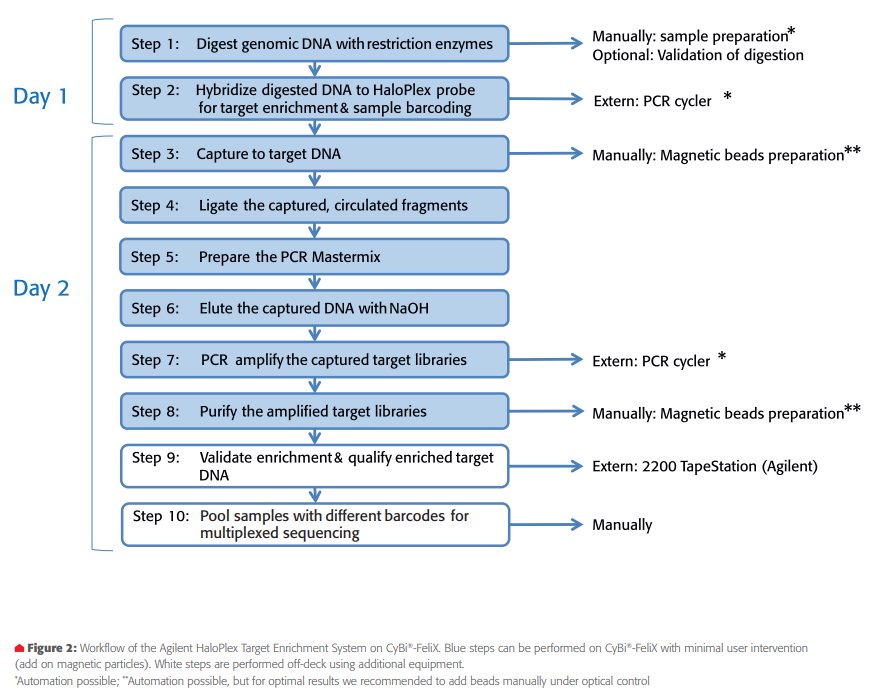
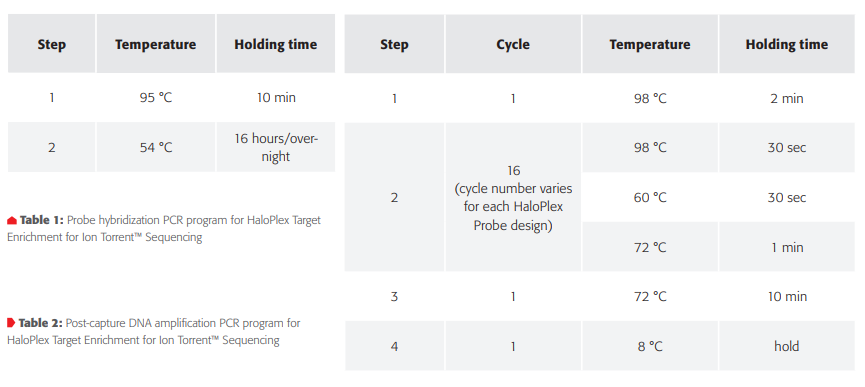
Experimental design & analysisAutomated library preparation was performed on the CyBi®-FeliX platform. The automation of the method follows the HaloPlex Target Enrichment System Kit protocol (Agilent) with a few modifications. During the process the PCR plate and tubes are not centrifuged and not cooled, beside the Enzyme Reaction Strips on position 8 of the deck layout day 1, which was placed on a precooled PCR rack. Short incubations were performed on deck and the PCR plate was not sealed during short incubation. The automation workflow is outlined in Figure 2. The method incorporates some stop points recommended in the protocol for manual steps.
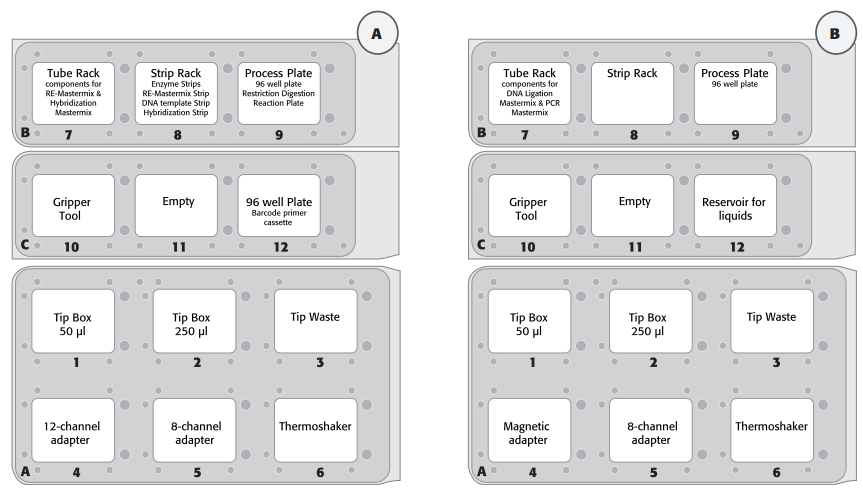
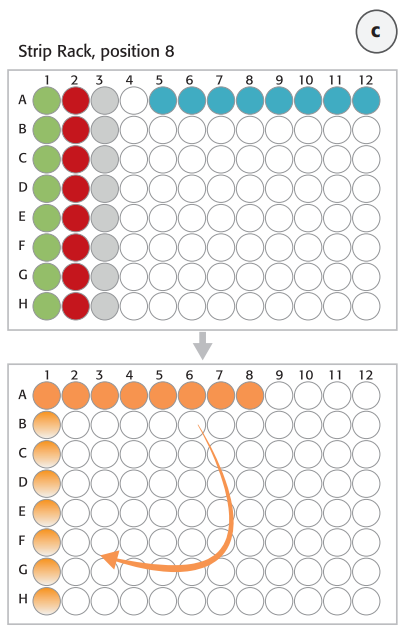
- The deck layout for the 2 day protocol is displayed in Figure 3. The deck layout was changed because of column by column and row wise distribution for digestion and hybridization on day 1 and magnetic bead-based capture and purification on day 2.
- To evaluate the automated procedure, 8 patient DNA sample libraries without an internal control were prepared using the CyBi®-FeliX workstation. Throughout the automated library preparation protocol, the bead solutions for capture target DNA (HaloPlex Magnetic Beads) and purification amplified target libraries (Agencourt AMPure XP beads) were prepared and added manually. Manual bead preparation is needed to prevent bead clumping which leads to a better homogeneous suspension and a uniform distribution of the beads on the PCR plate.
- The PCR library amplification was performed off-deck. Amplified (sequencing ready) libraries were analysed and quantified using a High Sensitivity D1000 ScreenTape with D1000 Reagents (both Agilent) for the Agilent 2200 TapeStation.
Detailed Protocol - Library PreparationDetailed guidelines for the construction of libraries with the HaloPlex Target Enrichment System Kit for Ion Torrent™ Sequencing on the CyBi®-FeliX are provided below. Please refer to the protocol for all instructions not included in this document. The following protocol includes volumes appropriate for 8 samples, 2 reaction excess. The protocol workflow with 2 different deck layouts (Figure 3) is spread on 2 days because of a 16 hour Hybridization step.
Step 1. Digest genomic DNA with restriction enzymes. Normalized genomic DNA (gDNA) samples were prepared and diluted manually and provided in an 8-well strip (Figure 3C, blue tube strip). The Restriction Enzyme Master Mix was prepared and the digestion process was performed on the thermal shaker (BioShake 3000 T elm) on deck. The restriction digestion reaction was validated by using the Agilent 2200 TapeStation.
Step 2. Hybridize digested DNA to HaloPlex probe for target enrichment and sample barcoding. HaloPlex probe was designed to hybridize selectively to target regions of the genome. 8-well strips from Step1 (Enzyme Strips, RE-Mastermix Strip) on position 8 (columns 1-3) were manually discarded and replaced by a new 8-well strip for the Hybridization Master Mix (Figure 3C, orange tube strip). The digested DNA samples from the 96-well Restriction Digest Reaction Plate were pooled with the 12-channel adapter in the hybridization reaction strip. After DNA pooling the hybridization reaction strip had to be manually rotated before adding the HaloPlex ION Barcode Primer Cassette to each tube using 8-channel liquid handling adapter (Figure 3C). The hybridization step was performed overnight external in a thermal cycler using the thermal cycler program from Table 1.
Step 3. Capture the target DNA. Manually resuspended and prepared Streptavidin[1]coated magnetic beads were provided and transferred manually. For optimal results we recommended adding beads under optical control to ensure that a homogenous bead suspension was distributed to all samples. The following mixing and washing steps were performed automatically.
Step 4. Ligate the captured, circularized fragments. Ligation Master Mix was prepared and added to the DNA captured reactions, followed by incubation in the on-deck thermal shaker.
Step 5. Prepare the PCR Master Mix. A PCR Master Mix was prepared for the captured target DNA amplification step, distributed to 0.2 ml 8-well strip.
Step 6. Elute captured DNA with NaOH. After ligation reaction the captured DNA libraries were eluted with freshly-prepared 50 mM NaOH.
Step 7. PCR amplify the captured target libraries. Libraries were amplified within an external thermal cycler with the PCR Master Mix prepared in step 5. Cycling was performed external in thermal cycler with parameters given in Table 2.
Step 8. Purify the amplified target libraries. Amplified DNA was purified using AMPure XP beads. The beads were prepared manually as recommended in the protocol and added manually to the samples as mentioned in step 3. Amplified libraries are eluted in 10 mM Tris-EDTA and transferred into a new column of the process plate on position 9.
Step 9. Validate enrichment and quantify enriched target DNA. Size distribution of the final libraries was confirmed external using a 2200 TapeStation (Agilent). Size distribution of libraries should be in the range of 150 to 550 bp.
Step 10. Pool samples with different barcodes for multiplexed sequencing. This step was done manually according to the recommendations from the Agilent protocol.
Results and discussion
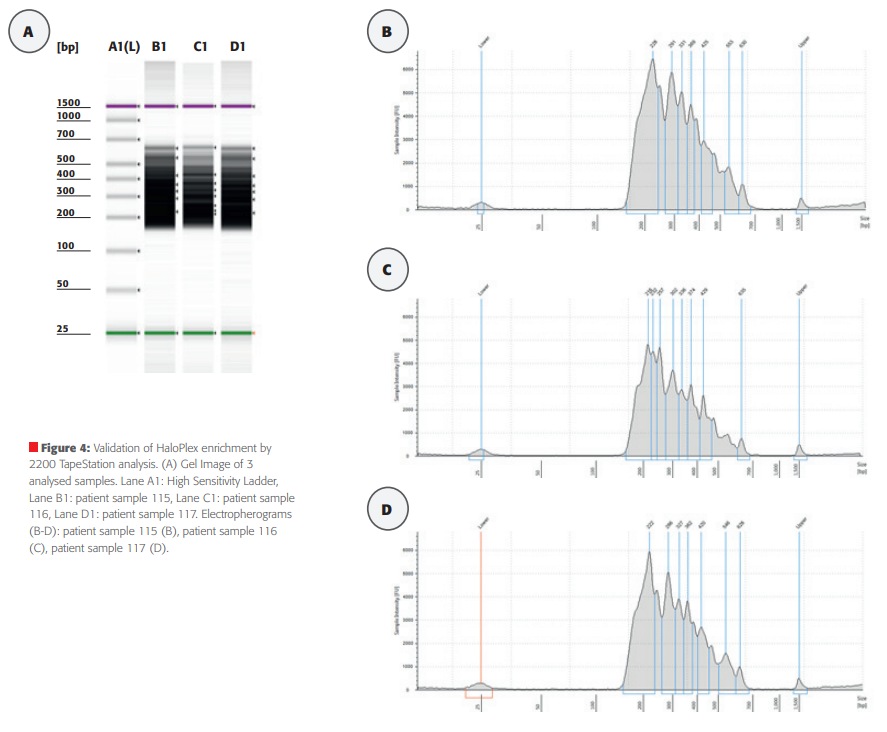
Sample analysis using the 2200 TapeSation 8 patient samples were used for automated library construction using HaloPlex Target Enrichment System Kit on the CyBi®-FeliX platform. To assess the quality of each library, the validation of enriched target DNA was analysed using the 2200 TapeStation (Agilent) according to the manufacturer´s instructions. If the library has been correctly assembled with adapters and barcodes to both ends of the fragment, there should be an electropherogram that shows a peak fragment size between approximately 150 to 550 bp. TapeStation data obtained from the analysis of 2 µl of each amplified libraries are shown in Figure 4. The gel image for 3 selected amplified libraries (patient sample 115 - 117) is presented in Figure 4A. All 3 samples prepared with the HaloPlex Target Enrichment System Kit on the CyBi®-FeliX were consistent and had the desired fragment size distribution, with a peak between 200 – 550 bp (Figure 4, B – D). The TapeStation-measured concentrations of the samples were approximately equal, 150.6 nmol/l for patient sample 115, 128.1 nmol/l for patient sample 116 and 126.3 nmol/l for patient sample 117. The yield was very good, only 100 pmol/l is for the sequencing process necessary. This data demonstrate the reliable automation of NGS library preparation with the CyBi®-FeliX.
Summary
Automated library sample preparation for Ion Torrent™ Sequencing can be successfully performed on the CyBi®-FeliX, using the HaloPlex Target Enrichment System Kit for Ion Torrent™ Sequencing. The proof of principle to construct 8 high-quality libraries in parallel was demonstrated. Moreover, the HaloPlex Target Enrichment System protocol from Agilent for Illumina Sequencing includes the same steps. The quality of the libraries prepared on the CyBi®-FeliX was comparable to that prepared manually. It should be noted that the use of CyBi®-FeliX for automation of NGS sample preparation using the HaloPlex Target Enrichment System Kit enables easy preparation of multiple samples with minimal effort and greater consistency. The CyBi®-FeliX is an ideal flexible liquid handling platform to improve reproducibility of the results and reduce the hands-on time.