Bruker’s very fast rapifleX MALDI PharmaPulse solution has been successfully used in pharmaceutical research for compound screening in various cell-free biochemical assays that measure distinct mass changes of synthetic or recombinant substrates that are affected by small molecule inhibitors.
Abstract
In this application note, we demonstrate a rapid cell-based assay with partly automated sample preparation for the profiling of BCR-ABL kinase inhibitors in K562 leukemia cells. Heme B, a marker for drug-induced redifferentiation of these cells and, hence, a suitable drug response marker, was used to establish concentration-response relationships for three drugs. The Bruker rapifleX MALDI PharmaPulse approach allowed for automated and label-free measurements of the response marker in treated cells at high speed.
Introduction
Matrix-assisted laser desorption/ ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS) is a versatile technology for the label-free analysis of various classes of molecules. Also in the field of compound screening in drug discovery, MALDI-TOF MS has become a powerful alternative approach due to recent technical advances in speed, automation and specificity of sample analysis. In contrast to common high throughput screening methods, there is no need for specific antibodies, probe development or labelling. Furthermore, typical pitfalls in fluorescence assays like autofluorescence or fluorescence quenching, can be avoided.
Recently, rapifleX MALDI PharmaPulse has successfully been used in various biochemical assays measuring mass changes upon compound treatment in pharmaceutical (incl. industrial) research [1-4]. In contrast, MALDI-MS-based cellular assays are still underexplored in pharmacology and chemical biology. Recent studies of whole mammalian cell analysis by MALDI-TOF MS [5,6] demonstrate that this technique can also be applied to monitor pharmacodynamics effects in cells. Concentrationdependent molecular changes can be measured for known and directly affected abundant proteins (e.g. polyacetylated histones) or low molecular mass molecules. More importantly, MALDI-TOF MS offers the opportunity to identify pharmacodynamic effects on a lipid or metabolic level by computational workflows, since hundreds of molecules can be scanned simultaneously. Here, we describe a fast and automated cellular phenotypic assay for heme inducers such as BCR-ABL inhibitors using the rapifleX MALDI PharmaPulse. This class of tyrosine kinase inhibitors blocks the detrimental function of the BCR-ABL fusion protein, the oncogenic driver in chronic myelogenous leukemia (CML) [7].

Methods
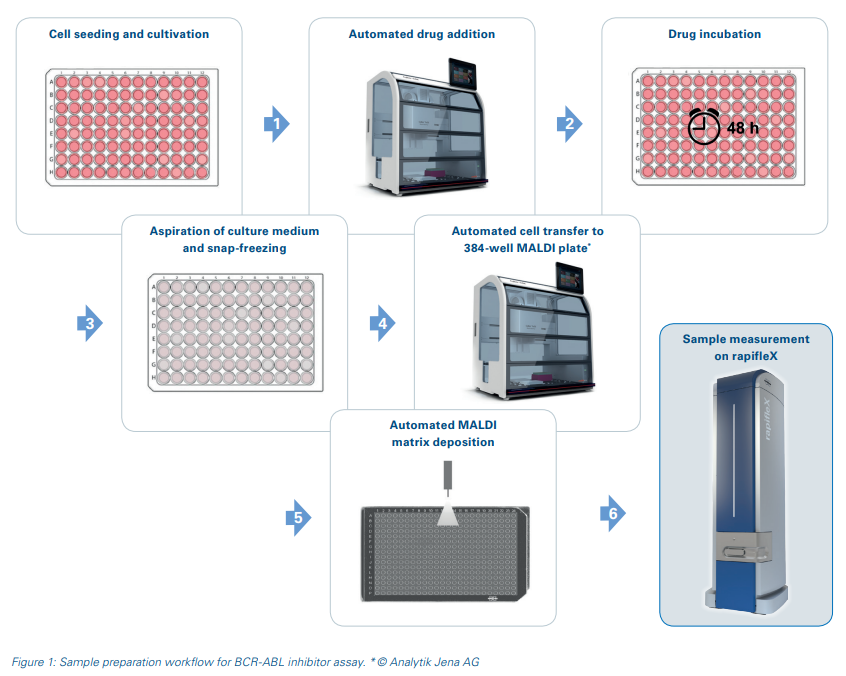
K562 cells were cultured in RPMI-1640 medium supplemented with 10% FCS, 1 mM sodium pyruvate and 2 mM L-Glutamine. For the drug response assay, cells were seeded in 96-well plates at a density of 0.1*106 cells per mL. After 16 hours, cells were treated with either dasatinib, imatinib, chloroquine or the vehicle control DMSO for 48 hours. Cells were then harvested by centrifugation at 800 rpm, aspiration of the supernatant and snap-freezing of the 96-well plate in liquid nitrogen. Cells were resuspended in 40 µL of water and two µL from each well were transferred to a MALDI steel target in 384-format in four technical replicates. For this study, four biological replicates were assessed.
Compound addition to the assay plate and transfer of resuspended treated cells to the MALDI target was performed by a CyBio FeliX automated liquid handling robot (Analytik Jena AG, Germany) to enhance assay reproducibility.
For MALDI-TOF MS, the target plate was spray-coated with DHB matrix (20 mg/mL in ACN/water (1:1 v/v) supplemented with 0.5% TFA) using a TM-sprayer (HTX Technologies LLC, Chapel Hill, NC, USA). The sample plate was subsequently measured on a rapifleX MALDI-TOF mass spectrometer (Bruker Daltonics). For detailed method parameters see Table 1.
Signal validation was performed on a solariX XR 7T MALDI magnetic resonance mass spectrometer (MRMS; Bruker Daltonics).
After recalibration in flexAnalysis 3.4, spectra were processed and analyzed as described previously [5]. Heme B signal intensities of technical replicates were averaged and the fold change to the vehicle control was calculated. The data was subsequently fitted to a non-linear regression dose-response model with variable slope using GraphPad Prism 5 (GraphPad Software, San Diego, USA).
Results and Discussion
BCR-ABL inhibitors are the first-line treatment of BCR-ABL-positive CML patients. In order to establish a MALDI-TOF-based inhibitor screen, we treated K562 leukemia cells with various concentrations of the two BCR-ABL inhibitors imatinib and dasatinib. The disease unrelated malaria inhibitor chloroquine was included as a control substance.
Cells are used directly from the cultivation plate for this MALDI-TOF assay and no sample cleanup steps are required. Drug addition to the cells and cell transfer to the MALDI target plates were carried out in an automated manner using the CyBio FeliX pipetting robot to ensure high reproducibility and to enable a higher sample throughput for potential drug screenings. After MALDI matrix application, the sample measurement was performed automatically on a rapifleX MALDI-TOF MS. The schematic workflow is displayed in Figure 1.
As candidate pharmacodynamic markers that could be measured in a cell assay, we identified concentration-dependent molecular changes at the lipid and metabolic level in response to treatment of K562 cells with the two BCR-ABL tyrosine kinase inhibitors but not with the unrelated compound.
The m/z value 616.2 was selected as a suitable response read-out although other m/z values showed equally a concentration-dependent change in signal intensity (see [5]). Using ultra-high resolving power MRMS (solariX, Bruker Daltonics), the response marker was identified as heme B. Heme B is a known marker for erythroid differentiation and therefore a functional measure of drug response in CML cells.
The specificity of the response signal was evaluated in the rapifleX MS spectra and additionally at higher mass resolution in the MRMS spectra measured on the same MALDI target plate (figure 2). No interfering signals were detected showing that the measured response was solely due to heme B induction.
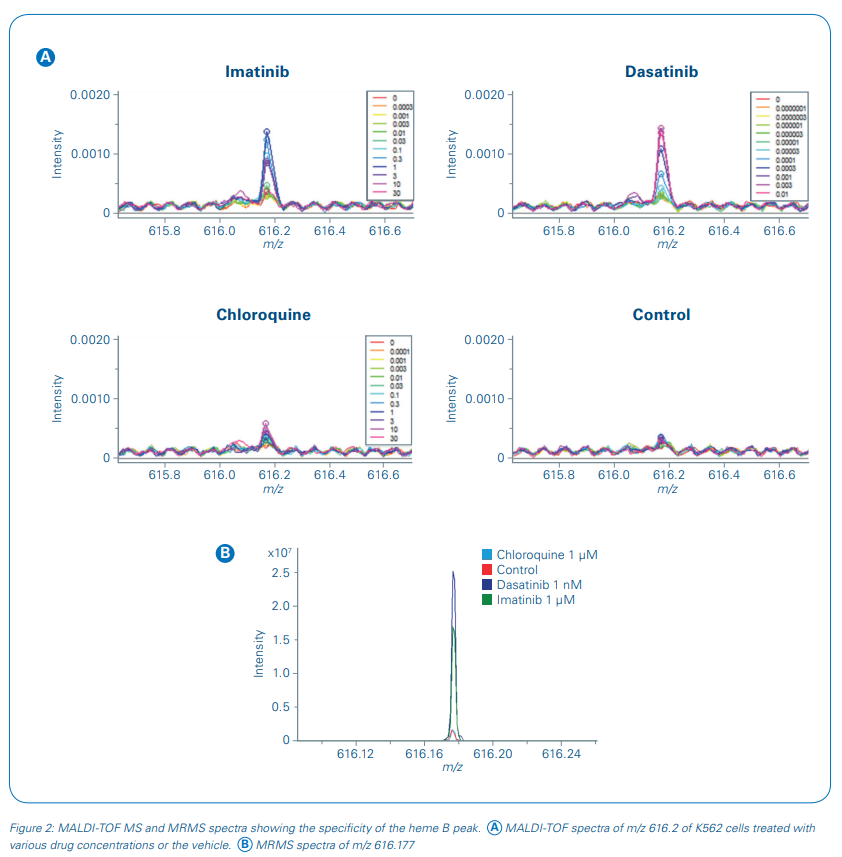
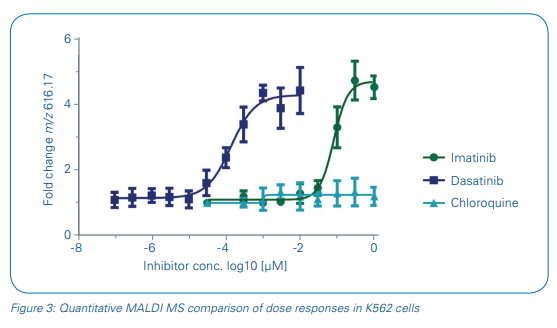
Consequently, we used heme B in this study to monitor the concentrationdependent pharmacodynamic effect directly in cells without further sample cleanup. Intensity values for m/z 616.2 were extracted and the fold change of the heme B signal intensity to the vehicle control intensity was applied for sigmoidal curve fitting and pIC50 determinations of the three test drugs (Figure 3). The pIC50 values of imatinib and dasatinib were calculated as 7.1 ± 0.1 and 9.8 ± 0.1, respectively, and showed a good reproducibility over the four biological replicates. pIC50s of our MALDI-TOF MS-based assay compared well to literature data [7,8].
Conclusions
- The metabolic response marker heme B was successfully applied as a concentrationdependent read-out for the quantitative measurement of drug inhibition in whole cells spotted onto a MALDI target plate. pIC50 values compared well to reference data from cell-based assays described in literature.
- MALDI-TOF MS in combination with a liquid handling robot enables automated, reproducible and label-free drug screening in whole cells without sample cleanup.
- The rapifleX MALDI-TOF MS allows for robust measurements at high speed